An Adaptive Genetic Algorithm with Recursive Feature Elimination Approach for Predicting Malaria Vector Gene Expression Data Classification using Support Vector Machine Kernels
DOI:
https://doi.org/10.48048/wjst.2021.9849Keywords:
RNA-seq, Adaptive genetic algorithm, Recursive feature elimination, Malaria vector, Support Vector Machine kernelsAbstract
As mosquito parasites breed across many parts of the sub-Saharan Africa part of the world, infected cells embrace an unpredictable and erratic life period. Millions of individual parasites have gene expressions. Ribonucleic acid sequencing (RNA-seq) is a popular transcriptional technique that has improved the detection of major genetic probes. The RNA-seq analysis generally requires computational improvements of machine learning techniques since it computes interpretations of gene expressions. For this study, an adaptive genetic algorithm (A-GA) with recursive feature elimination (RFE) (A-GA-RFE) feature selection algorithms was utilized to detect important information from a high-dimensional gene expression malaria vector RNA-seq dataset. Support Vector Machine (SVM) kernels were used as the classification algorithms to evaluate its predictive performances. The feasibility of this study was confirmed by using an RNA-seq dataset from the mosquito Anopheles gambiae. The technique results in related performance had 98.3 and 96.7 % accuracy rates, respectively.
HIGHLIGHTS
- Dimensionality reduction method based of feature selection
- Classification using Support vector machine
- Classification of malaria vector dataset using an adaptive GA-RFE-SVM
GRAPHICAL ABSTRACT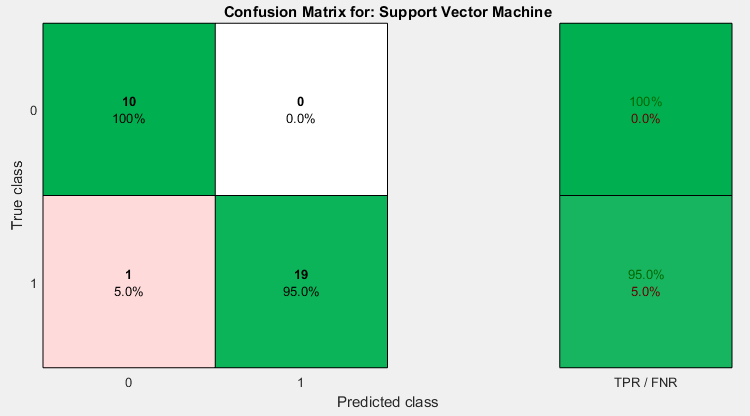
Downloads
Metrics
References
S Sun, C Wang, H Ding and Q Zou. Machine learning and its applications in plant molecular studies. Briefings Funct. Genom. 2019; 19, 40-8.
DF Read, K Cook, YY Lu, KGL Roch and WS Noble. Predicting gene expression in the human malaria parasite plasmodium falciparum using histone modification, nucleosome positioning, and 3D localization features. PLoS Comput. Biol. 2019; 15, e1007329.
MO Arowolo, M Adebiyi and A Adebiyi. A dimensional reduced model for the classification of RNA-seq Anopheles gambiae data. J. Theor. Appl. Inform. Tech. 2019; 97, 3487-96.
S Karthik, and M Sudha. A survey on machine learning approaches in gene expression classification in modelling computational diagnostic system for complex diseases. Int. J. Eng. Adv. Tech. 2018; 8, 182-91.
NT Johnson, A Dhroso, KJ Hughes and D Korkin. Biological classification with RNA-seq data: Can alternatively spliced transcript expression enhance machine learning classifiers? RNA 2018; 24, 1119-32.
MW Libbrecht and WS Noble. Machine learning applications in genetics and genomics. Nat Rev Genet. 2015; 16, 321-32.
Z Jagga and D Gupta. Classification models for clear cell renal carcinoma stage progression, based on tumor RNAseq expression trained supervised machine learning algorithms. BMC Proc. 2014; 8, S2.
The Anopheles gambiae 1000 Genomes Consortium. Genetic diversity of the African genetic diversity of the African malaria vector Anopheles gambiae. Nature 2017; 552, 96-100.
DH Oh, IB Kim, SH Kim and DH Ahn. Predicting autism spectrum disorder using blood-based gene expression signatures and machine learning. Clin. Psychopharmacol. Neurosci. 2017; 15, 47-52.
R Qi, A Ma, Q Ma and Q Zou. Clustering and classification methods for single-cell RNA-seq data. Brief. Bioinform. 2020; 21, 1196-208.
S Wenric and R Shemirani. Using supervised learning methods for gene selection in RNA-seq case-control studies. Front. Genet. 2018; 9, 1-6.
J Alquicira-Hernandez, A Sathe, HP Ji, Q Nquyen and JE Powell. scPred: Accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 2019; 20, 264.
S Cui, Q Wu, J West and J Bai. Machine learning-based microarray analyses indicate low-expression genes might collectively influence PAH disease. PLoS Comput. Biol. 2019; 15, e1007264.
HS Shon, YG Yi, KO Kim, EJ Cha and KA Kim. Classification of stomach canacer gene expression data using CNN algorithm of deep learning. J. Biomed. Transl. Res. 2019; 20, 15-20.
AJ Reid, AM Talman, HM Bennett, AR Gomes, MJ Sanders, CJR Illingworth, O Billker, M Berriman and MKN Lawniczak. Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. Elife 2018; 7, e33105.
AC Tan and D Gilbert. Ensemble machine learning on gene expression data for cancer classification. Appl. Bioinformatics 2003; 2, S75-S83.
N Song, K Wang, M Xu, X Xie, G Chen and Y Wang. Design and analysis of ensemble classifier for gene expression data of cancer. Adv. Genet. Eng. 2016; 5, 1000152.
S Tarek, RA Elwahab and M Shoman. Gene expression based cancer classification. Egypt. Informat. J. 2017; 18, 151-9.
M Bonizzoni, E Ochomo, WA Dunn, M Britton, Y Afrane, G Zhou, J Hartsel, MC Lee, J Xu, A Githeko, J Fass and G Yan. RNA-seq analyses of changes in the Anopheles gambiae transcriptome associated with resistance to pyrethroids in Kenya: Identification of candidate-resistance genes and candidate-resistance SNPs. Parasites Vector 2015; 8, 474.
G James, D Witten, T Hastie and R Tibshirani. An introduction to statistical learning: With application in R. Springer, New York, 2013.
B Duval and JK Hao. Advances in metaheuristics for gene selectio and classification of microarray data. Brief. Bioinform. 2010; 11, 127-41.
AK Shukla, P Singh and M Vardhan. A new hybrid feature subset selection framework based on binary genetic algorithm and information theory. Int. J. Comput. Intell. Appl. 2019; 18, 1950020.
AC Tan and D Gilbert. Ensemble machine learning on gene expression data for cancer classification. Appl. Bioinformatics 2003; 3, S57-83.
K Kowsari, KJ Meimandi, M Heidarysafa, S Mendu, LE Barnes and DE Brown. Text classification algorithms: A survey. Information 2019; 10, 150.
AM Olaolu, SO Abdulsalam, IR Mope and GA Kazeem. A comparative analysis of feature selection and feature extraction models for classifying microarray dataset. Comput. Inform. Syst. 2018; 22, 29-38.
H Aydadenta and Adiwijaya. On the classification techniques in data mining for microarray data classification. J. Phys. Conf. Series. 2018; 971, 012004.
CC Chang and CJ Lin. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Tech. 2011; 2, 27.
A. Khan, B. Baharudin, L.H. Lee, K. khan, K. A Review of Machine Learning Algorithms for Text-Documents Classification. Journal of Advances in Information Technology. 2010: 1; pp. 1-17.
HP Bhavsar and M Panchal. A review on support vector machine for data classification. Int. J. Adv. Res. Comput. Eng. Tech. 2012; 1, 185-9.
CDA Vanitha. D Devaraj and M Venkatesulu. Gene expression data classification using support vector machine and mutual information-based gene selection. Proc. Comput. Sci. 2015; 47; 13-21.
MO Arowolo, SO Abdulsalam, RM Isiaka and KA Gbolagade. A hybrid dimensionality reduction model for classification of microarray dataset. Int. J. Inform. Tech. Comput. Sci. 2017; 9, 57-63.
AK Shukla. Multi-population adaptive genetic algorithm for selection of microarray biomarkers. Neural Comput. Appl. 2020; 32, 11897-918.
XW Chen and JC Jeong. Enhanced recursive feature elimination. In: Proceedings of the 6th International Conference on Machine Learning and Applications, Cincinnati, OH, USA. 2007, p. 429-35.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2021 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.