Cloning, Expression and Structural Modeling of the MlrA Protein from Novosphingobium sp. KKU25s for Microcystin Degradation
DOI:
https://doi.org/10.48048/wjst.2021.9455Keywords:
Microcystin, MlrA protein, Microcystinase, Cloning, Expression, Structural modelingAbstract
MlrA is a gene involved in the degradation of toxic cyanobacterial microcystins. This gene encodes microcystinase, mlrA, the 1st enzyme in the pathway that breaks down toxic cyanobacterial microcystins. In this study, primers were designed, and polymerase chain reaction (PCR) was performed to amplify the mlrA gene in Novosphincgobium sp. KKU25s. A PCR product of 752 base pairs was obtained. The nucleotide sequence of the mlrA gene of Novosphingobium sp. KKU25s was similar to that of Sphingomonas sp. ACM-3962 (98 % similarity). The mlrA gene of Novosphingobium sp. KKU25s was further cloned into the pGEM T-Easy plasmid to obtain the nucleotide sequence of the mlrA gene. The gene was also ligated into the pET32a plasmid for gene expression. Expression was induced by isopropyl β-D-1-thiogalactopyranoside (IPTG) and verified using SDS-PAGE. The expressed protein was approximately 22 kilodaltons. The cell-free extract (CE) containing the crude protein from confirmed recombinant cells showed high activity in the biodegradation of [Dha7] MC-LR. [Dha7] MC-LR at an initial concentration of 30 mg L-1 and was completely biodegraded within 30 h. A distinct product derived from [Dha7] MC-LR appeared with a decrease in the [Dha7] MC-LR peak in the HPLC profile. The product (m/z 999.51) showed a molecular weight of 18, which is higher than that of native [Dha7] MC-LR (m/z 981.50), and was determined to be a linearized peptide fragment of [Dha7] MC-LR using LC-MS analysis. The 3-dimensional structure of microcystinase was predicted from the amino acid sequence deduced from the mlrA gene by the Swiss Model and Phyre2 programs. The structure contained a predicted alpha helix. The predicted 3-dimensional structure was also similar to that of a protein in the CAAX protease group.
HIGHLIGHTS
- Research focused on characterization of microcystinase (MlrA) protein
- First research worked on the degradation of [Dha7] MC-LR by MlrA
- This work is useful for the applications aimed at the removal of MCs in freshwater environments
GRAPHICAL ABSTRACT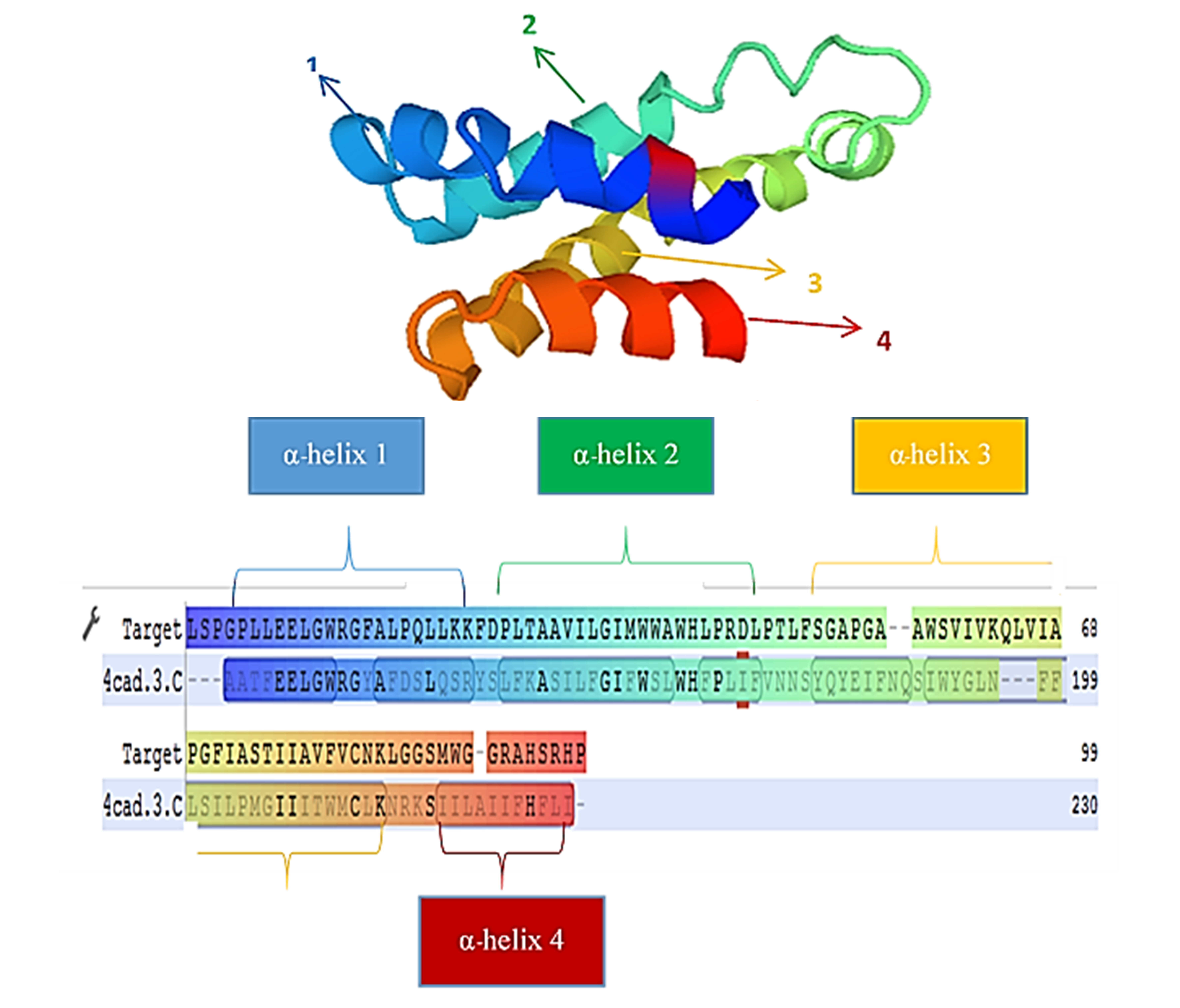
Downloads
Metrics
References
A Zyska and J Jasik-Ślęza. Mechanism and effects of cyanobacterial hepatotoxin action on human organism. Pol. J. Public Health 2014; 124, 156-9.
N Butler, JC Carlisle, R Linville and B Washburn. Microcystins: A brief overview of their toxicity and effects, with special reference to fish, wildlife, and livestock. California Environmental Protection Agency, Sacramento, USA, 2009, p. 1-17.
M Rex. Is protein phosphatase inhibition responsible for the toxic effects of okadaic acid in animals? Toxins 2013; 2, 267-85.
I Trevino-Garrison, J DeMent, FS Ahmed, P Haines-Lieber, T Langer, H Ménager, J Neff, van der D Merwe and E Carney. Human illnesses and animal deaths associated with freshwater harmful algal blooms-Kansas. Toxins 2015; 2, 353-66.
RP Rastogi, RP Sinha and A Incharoensakdi. The cyanotoxin-microcystins: Current overview. Rev. Environ. Sci. Biotechnol. 2014; 13, 215-49.
RW Zurawell, H Chen, JM Burke and EE Prepas. Hepatotoxic cyanobacteria: A review of the biological importance of microcystins in freshwater environments. J. Toxicol. Environ. Health B Crit. Rev. 2005; 8, 1-37.
SJ Hoeger, BC Hitzfeld and DR Dietrich. Occurrence and elimination of cyanobacterial toxins in drinking water treatment plants. Toxicol. Appl. Pharmacol. 2005; 203, 231-42.
L Shang, M Feng, X Xu, F Liu, F Ke and W Li. Co-occurrence of microcystins and taste-and-odor compounds in drinking water source and their removal in a full-scale drinking water treatment plant. Toxins 2018; 1, 1-17.
T Somdee, J Wibuloutai, TD Somdee and A Somdee. Biodegradation of the cyanobacterial hepatotoxin [Dha7] MC-LR within a biologically active sand filter. Water Sci. Technol. Water Supply 2014; 4, 672-80.
Y Phujomjai, A Somdee and T Somdee. Biodegradation of microcystin [Dha7] MC-LR by a novel microcystin-degrading bacterium in an internal airlift loop bioreactor. Water Sci. Tech. 2016; 73, 267-74.
K Christoffersen, S Lyck and A Winding. Microbial activity and bacterial community structure during degradation of microcystins. Aquat. Microb. Ecol. 2002; 27, 125-36.
J Li, R Li and J Li. Current research scenario for microcystins biodegradation: A review on fundamental knowledge, application prospects and challenges. Sci. Total. Environ. 2017; 595, 615-32.
P Kumar, K Hedge, SK Brar, M Cledon and A Kermanshahi-pour. Potential of biological approaches for cyanotoxin removal from drinking water: A review. Ecotox. Environ. Saf. 2019; 172, 488-503.
DG Bourne, RL Blakeley, GJ Jones, A Jones, AP Negri and P Riddles. Enzymatic pathway for the acterial degradation of cyanobacterial cyclic peptide toxin microcystin-LR. Appl.Environ. Microbiol. 1996; 62, 4086-94.
DG Bourne, W Smith, GJ Jones and P Riddles. Characterization of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin-LR. Environ. Toxicol. 2001; 16, 523-34.
Y Peerapornpisal, W Sonthichai, M Suchotiratana, S Lipigorngoson, W Ruangyuttikarn, K Ruangrit, J Pekkoh, R Prommana, N Panuvanitchakorn, N Ngearnpat, S Kiatpradub and S Promkutkaew. Survey and monitoring of toxic cyanobacteria in water supplied and fisheries in Thailand. Chiang Mai J. Sci. 2002; 29, 71-9.
R Prommana, Y Peerapornpisal, N Whangchai, LF Morrison, JS Metcalf, W Ruangyuttikarn, A Towproma and GA Codd. Microcystins in cyanobacterial blooms from two freshwater prawn (Macrobrachium rosenbergii) ponds in Northern Thailand. Sci. Asia 2006; 32, 365-70.
T Somdee, T Kaewsan and A Somdee. Monitoring toxic cyanobacteria and cyanotoxins (microcystins and cylindrospermopsins) in four recreational reservoirs (Khon Kaen, Thailand). Environ. Monit. Assess. 2013; 185, 9521-9.
S Ruangsomboon, W Yongmanitchai, P Taveekijakarn and M Ganmanee. Cyanobacterial composition and microcystin accumulation in catfish pond. Chiang Mai J. Sci. 2014; 41, 27-38.
K Moolwangand and T Somdee. Purification of cyanobacterial toxin, microcystins, by DEAE and Strata-X SPE chromatography. KKU Sci. J. 2019; 47, 26-33.
EML Janssen. Cyanobacterial peptides beyond microcystins: A review on co-occurrence, toxicity, and challenges for risk assessment. Water Res. 2019; 151, 488-99.
JJ Doyle and JL Doyle. Isolation of plant DNA from fresh tissue. Focus 1990; 12, 13-5.
YF Cheng, CH Yang and WH Liu. Cloning and expression of Thermobifida xylanase gene in the methylotrophic yeast Pichia pastoris. Enzyme Microb. Technol. 2005; 37, 541-6.
J Sambrook and DW Russell. Purification of nucleic acids by extraction with phenol: Chloroform. CSH Protoc. 2006; 1, 169-70.
H Xu, H Wang, Q Xu, L Lv, C Yin, X Liu, H Du and H Yan. Pathway for biodegrading microcystin-YR by Sphingopyxis sp. USTB-05. PLoS One 2015; 10, e0124425.
J Dexter, D Dziga, J Lv, J Zhu, W Strzalka, A Maksylewicz, M Maroszek, S Marek and P Fu. Heterologous expression of mlrA in a photoautotrophic host-engineering cyanobacteria to degrade microcystins. Environ. Pollut. 2018; 237, 926-35.
D Dziga, M Wasylewski, B Wladyka, S Nybom and J Meriluoto. Microbial degradation of microcystins. Chem. Res. Toxicol. 2013; 26, 841-52.
R Wang, J Li, Y Jiang, Z Lu, R Li and J Li. Heterologous expression of mlrA gene originated from
Novosphingobium sp. THN1 to degrade microcystin-RR and identify the first step involved in degradation pathway. Chemosphere 2017; 184, 159-67.
T Saito, K Okano, HD Park, T Itayama, Y Inamori, BA Neilan, BP Burns and N Sugiura. Detection and sequencing of the microcystin LR-degrading gene, mlrA, from new bacteria isolated from Japanese lakes. FEMS Microbiol. Lett. 2003; 229, 271-6.
H Yan, W Wei, J Chen, J Wang and H Wang. Characterization of the first step involved in enzymatic
pathway for microcystin-RR biodegraded by Sphingopyxis sp. USTB-05. Chemosphere 2012; 87, 12-8.
D Dziga, B Wladyka, G Zielinska, J Meriluoto and M Wasylewski. Heterologous expression and characterization of microcystinase. Toxicon 2012; 59, 578-86.
K Tsuji, M Asakawa, Y Anzai, T Sumino and KI Harada. Degradation of microcystins using immobilized microorganism isolated in an eutrophic lake. Chemosphere 2006; 65, 117-24.
H Wang, L Ho, DM Lewis, JD Brookes and G Newcombe. Discriminating and assessing adsorption and biodegradation removal mechanisms during granular activated carbon filtration of microcystin toxins. Water Res. 2007; 41, 4262-70.
L Ho, D Hoefel, CP Saint and G Newcombe. Isolation and identification of a novel microcystin degrading bacterium from a biological sand filter. Water Res. 2007; 41, 4685-46.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2021 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.